This is the Linux app named AmPEP and AxPEP whose latest release can be downloaded as deep-ampep30-test_set.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named AmPEP and AxPEP with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
AmPEP and AxPEP
DESCRIPTION
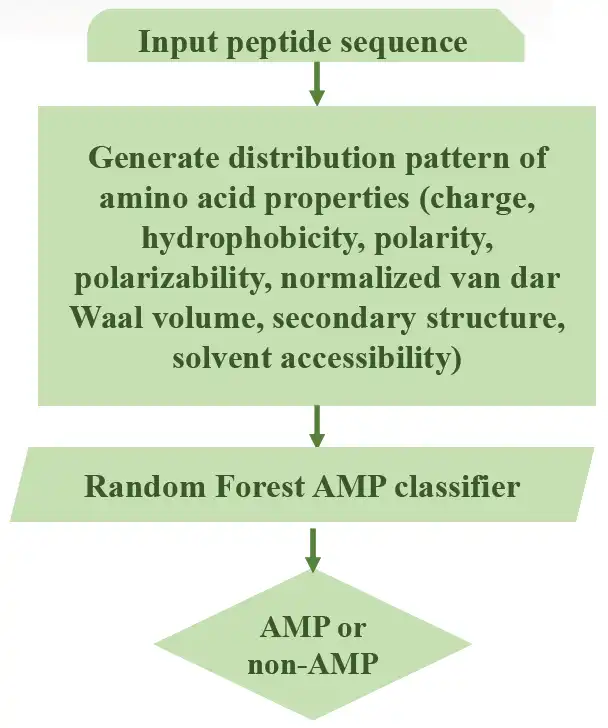
Antimicrobial peptides (AMPs) are promising candidates in the fight against multidrug-resistant pathogens due to its broad range of activities and low toxicity. However, identification of AMPs through wet-lab experiment is still expensive and time consuming. AmPEP is an accurate computational method for AMP prediction using the random forest algorithm. The prediction model is based on the distribution patterns of amino acid properties along the sequence. Our optimal model, AmPEP with 1:3 data ratio achieved a very high accuracy of 96%, MCC of 0.9, AUC-ROC of 0.99 and Kappa statistic of 0.9. AmPEP outperforms existing methods with respect to accuracy, MCC, and AUC-ROC when tested using the benchmark datasets.Features
- MATLAB source code
- AmPEP datasets
- Machine learning
- Bioinformatics
- Peptide sequence
- Drug discovery
This is an application that can also be fetched from https://sourceforge.net/projects/axpep/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.