This is the Linux app named CIMS to run in Linux online whose latest release can be downloaded as CIMS.v1.0.5.tgz. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named CIMS to run in Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
CIMS to run in Linux online
DESCRIPTION
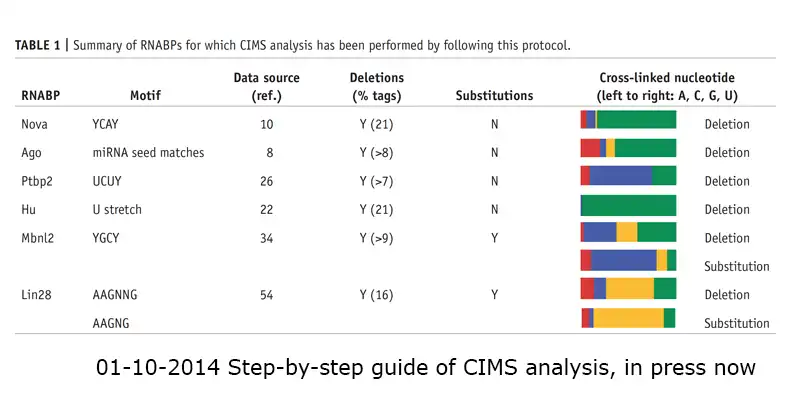
This package includes the scripts to detect statistically reproducible crosslinking induced mutation sites (CIMS) and cross linking induced truncation sites (CITS) from HITS-CLIP data.References:
Moore, M.*, Zhang, C.*, Gantman, E.C., Mele, A., Darnell, J.C., Darnell, R.B. 2014. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protocols, 9:263-293.
Zhang,C.†, Darnell, R.B.† 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotech. 29:607-614.
Features
- Mapping raw CLIP reads to the reference genome (using novoalign)
- Collapsing PCR duplicates with different models depending on presence of random barcodes
- Cluster CLIP tags to identify clusters and determine peak heights
- Tracking mutations (insertions, deletions and substitutions) in unique CLIP tags
- Identifying robust cross linking induced mutation sites (CIMS)
- Identifying robust cross linking induced truncation sites (CITS) from BrdU-CLIP, iCLIP or other similar variations
Audience
Advanced End Users
User interface
Command-line
Programming Language
Perl
This is an application that can also be fetched from https://sourceforge.net/projects/ngs-cims/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.