This is the Linux app named dataMAPPs to run in Linux online whose latest release can be downloaded as dataMAPPs_3.2.7.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named dataMAPPs to run in Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
dataMAPPs to run in Linux online
DESCRIPTION
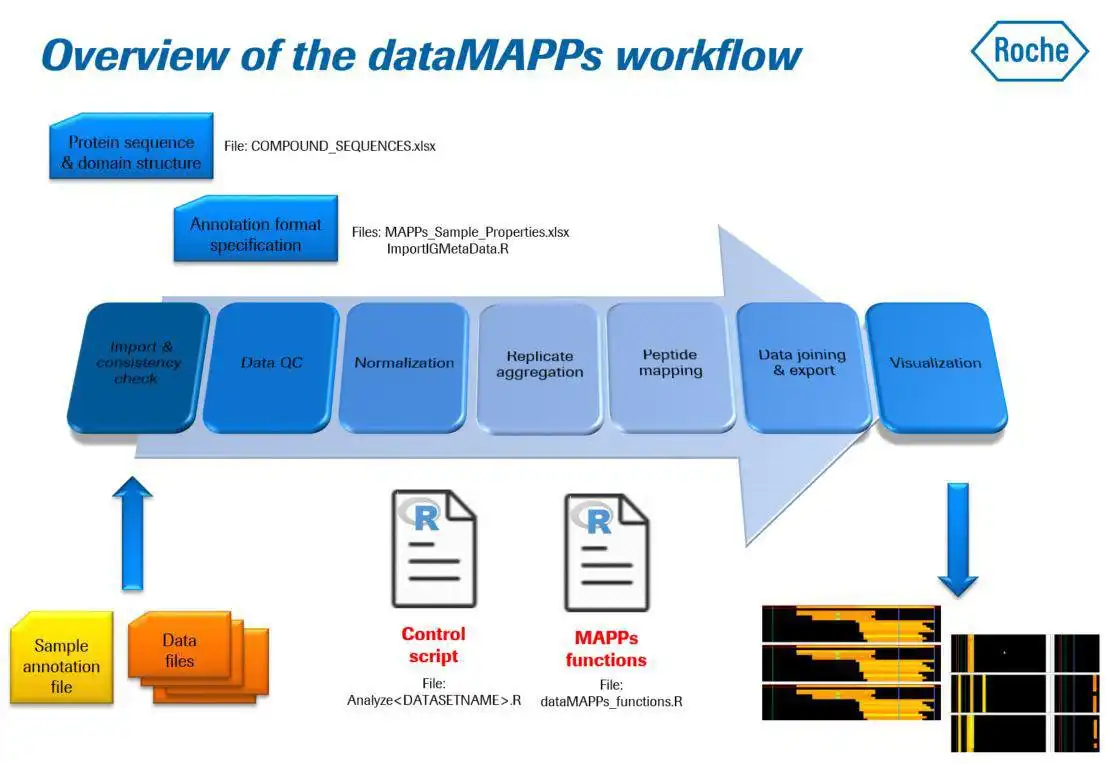
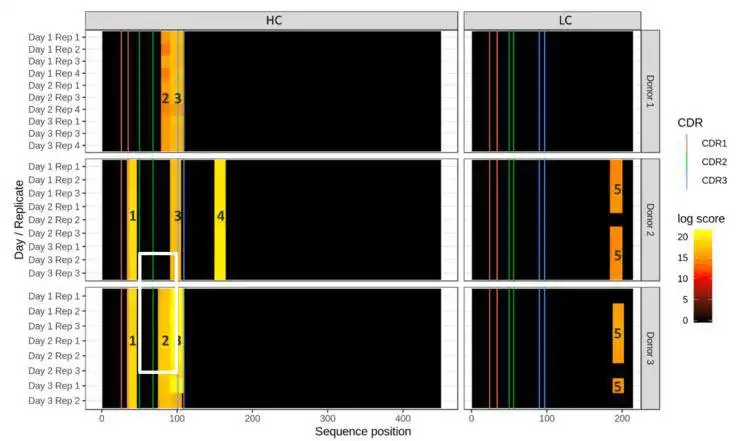
dataMAPPs allows routine and efficient processing of data from immunogenicity studies applying the MAPPs peptidomics technology to detect potential MHCI- or MHC-II epitopes as presented by dendritic cells (DC).It features quality control of the raw data, across-sample/across-donor normalization and visualization of results in a heatmap style (heatMAPPs). dataMAPPs' core is a generic R library that can be tailored to specific projects via dedicated control scripts which also allow reproducible recalculation of results.
Consult README file for installation and usage instructions. Further documentation is supplied in PDF format.
dataMAPPs is published under GPL license by a team of scientists working for Hoffmann-La Roche AG in Basel, Switzerland. The software is provided as is, to hopefully benefit other researchers.
Publication in preparation:
Steiner et al, 'Enabling Routine MHC-II Associated Peptide ProteomicS (MAPPs) for Risk Assessment of Drug-Induced Immunogenicity'
Features
- import of peptidomics data (PEAKS output, but may be extended)
- extensive data QC and sample exclusion by user definable cutoffs
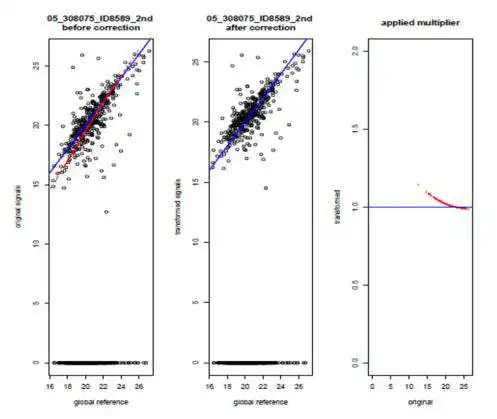
- dedicated 2-step normalization procedure for MAPPs data
- mapping peptides to target antibody/protein
- clustering peptide mapping positions into hotspot epitope regions
- visualization of mapping results as heatmaps
Audience
Science/Research
User interface
Console/Terminal
Programming Language
S/R
Database Environment
Flat-file
This is an application that can also be fetched from https://sourceforge.net/projects/datamapps/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.