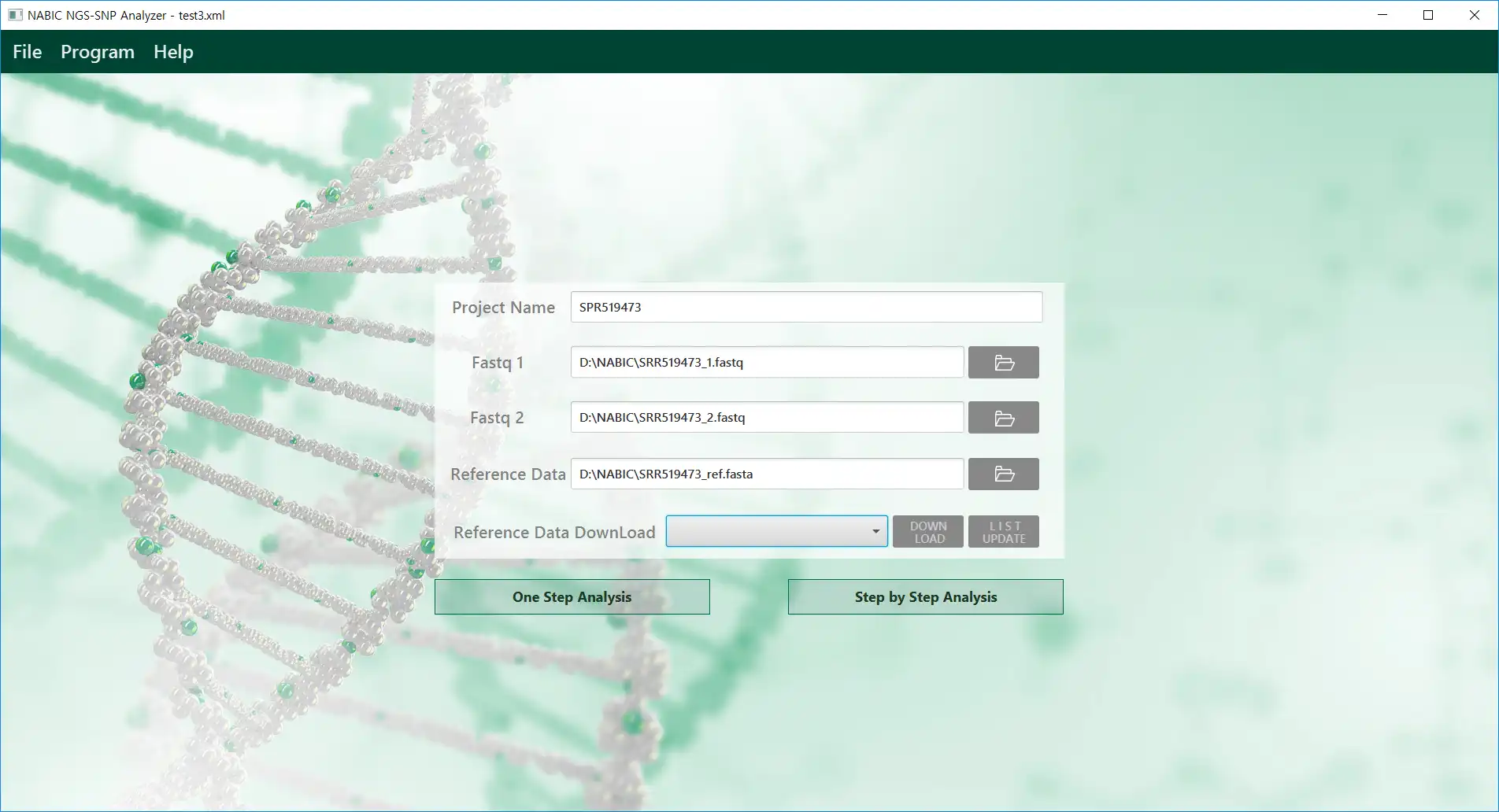
This is the Linux app named NGS_SNPAnalyzer to run in Linux online whose latest release can be downloaded as dataset.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named NGS_SNPAnalyzer to run in Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
NGS_SNPAnalyzer to run in Linux online
DESCRIPTION
NGS_SNPAnalyzer: A desktop software and fully automated graphical pipeline. The NGS_SNPAnalyzer not only includes the functionalities for variant calling and annotation but also provides details of quality control, mapping, and filtering to support all the procedures from next-generation sequencing (NGS) sequencing data to variant visualizationAudience
Science/Research
User interface
JavaFX
Programming Language
Java
Database Environment
XML-based
This is an application that can also be fetched from https://sourceforge.net/projects/ngs-snpanalyzer/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.