This is the Linux app named NGSEP to run in Linux online whose latest release can be downloaded as NGSEPcore_4.0.1.jar. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named NGSEP to run in Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
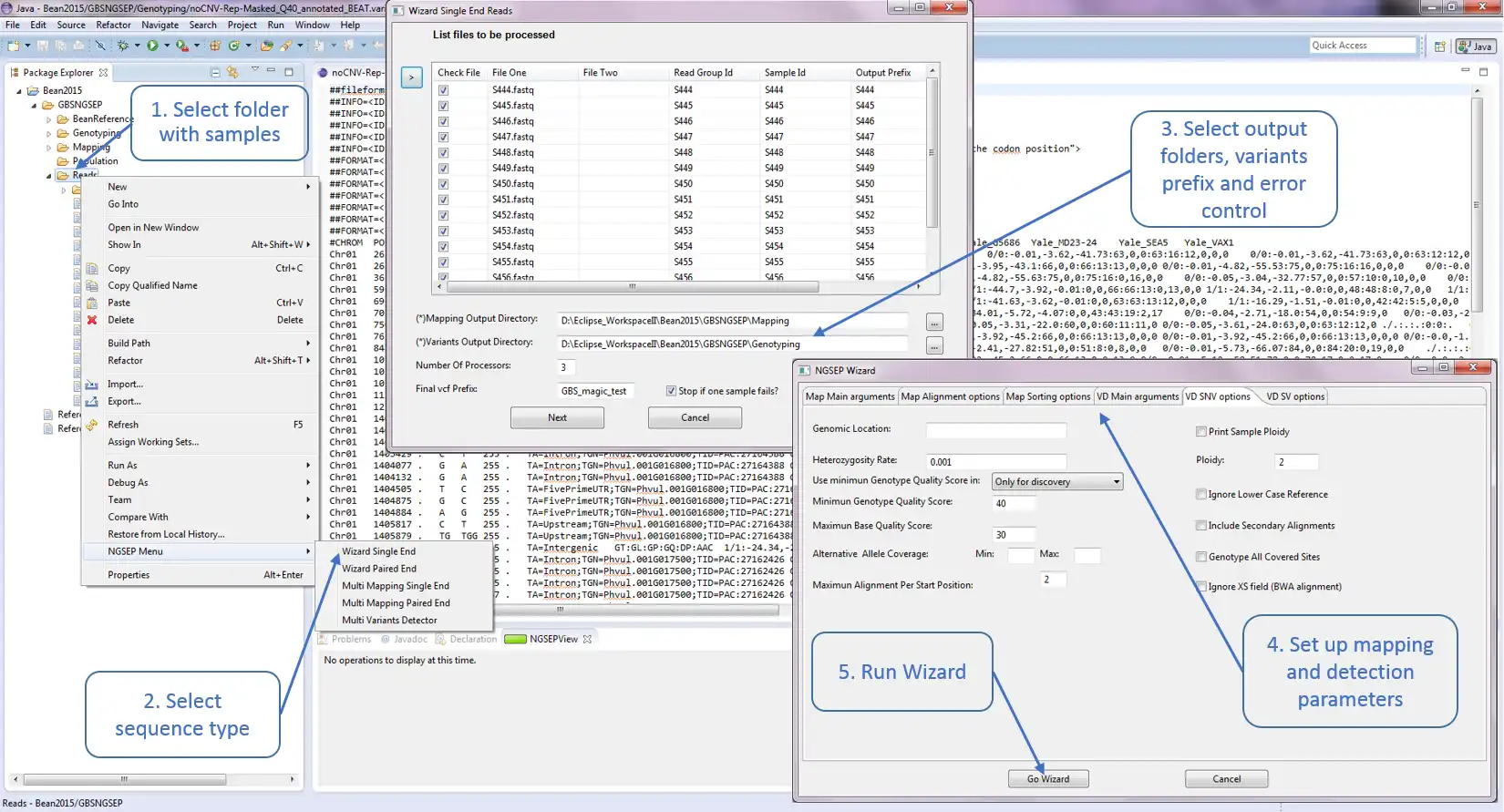
SCREENSHOTS
Ad
NGSEP to run in Linux online
DESCRIPTION
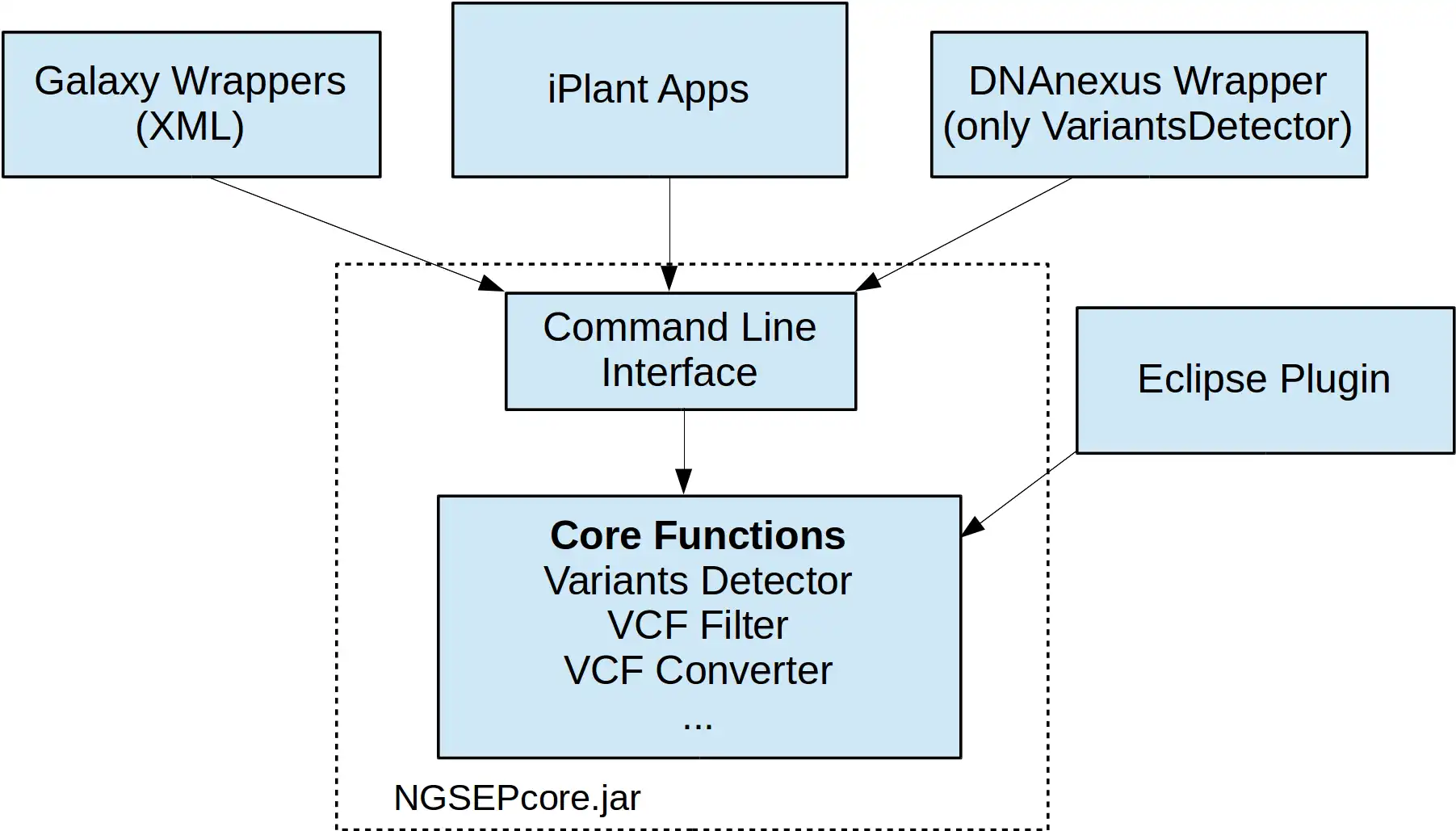
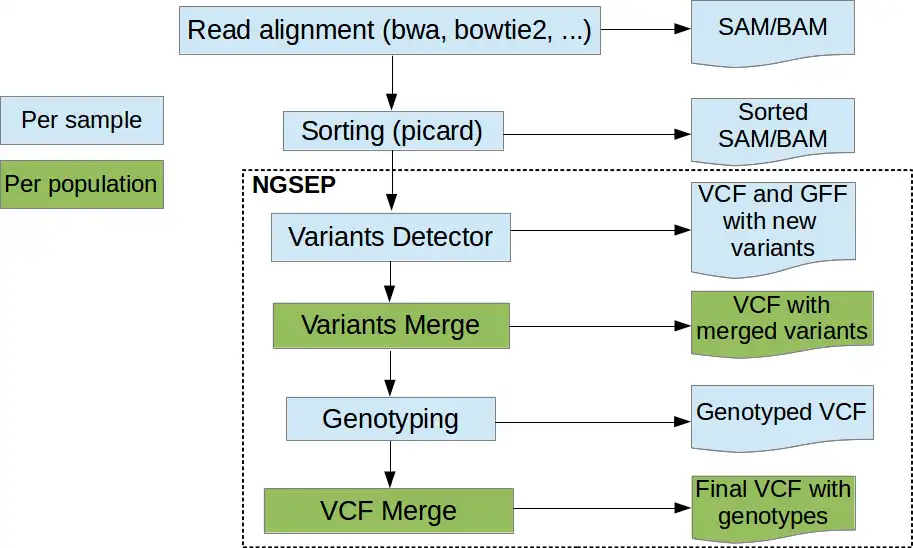
NGSEP is an integrated framework for analysis of DNA high throughput sequencing data. The main use of NGSEP is the construction and downstream analysis of large datasets of genomic variation. NGSEP performs accurate detection and genotyping of Single Nucleotide Variants (SNVs), small and large indels, short tandem repeats (STRs), inversions, and Copy Number Variants (CNVs). NGSEP also provides modules for functional annotation, filtering, format conversion, comparison, clustering, imputation, introgression analysis and different kinds of statistics. A complete list of functionalities is available in our wiki (https://sourceforge.net/p/ngsep/wiki/Home/).NEWS: We just made available the first command line release of version 4, including our own implementation of an FM-index based reads aligner. The command line usage was standardized over the entire application. Please take time to update scripts to upgrade to this new version. Further details are available in the wiki.
Features
- SNPs, CNVs and Structural Variants detection
- Alignment of raw reads to a reference genome
- VCF manipulation: functional annotation, merge, filter, compare, format conversion, imputation
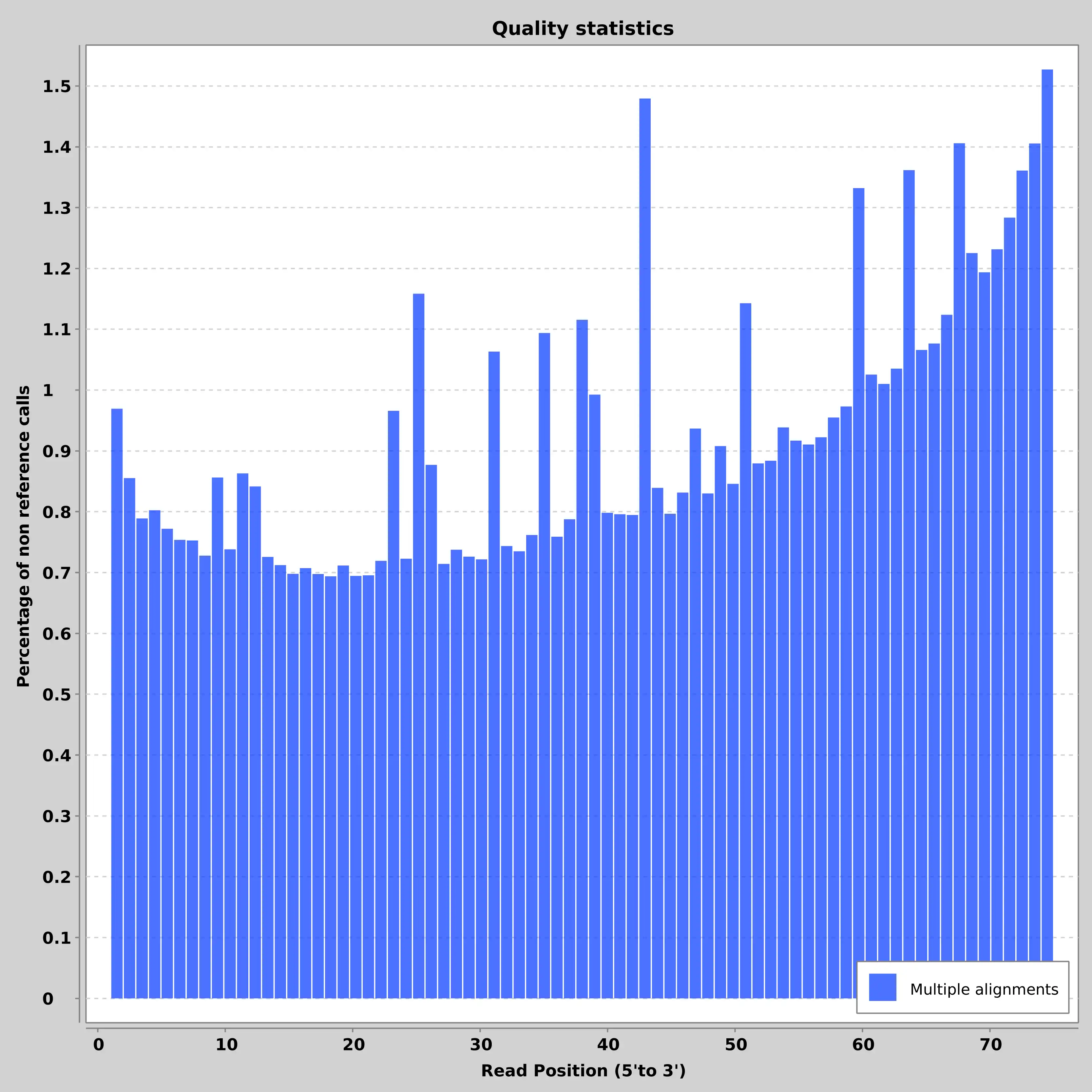
- SAM/BAM and VCF statistics calculation and plotting
- Reads Demultiplexing
- Alignment of annotated genome assemblies
- Statistics and filtering on transcriptome annptations in GFF3 format
Audience
Healthcare Industry, Science/Research, Other Audience, Agriculture
User interface
Java SWT, Console/Terminal, Eclipse
Programming Language
Java
This is an application that can also be fetched from https://sourceforge.net/projects/ngsep/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.