This is the Windows app named Calis-p to run in Windows online over Linux online whose latest release can be downloaded as calis-p-0.1.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named Calis-p to run in Windows online over Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
Calis-p to run in Windows online over Linux online
DESCRIPTION
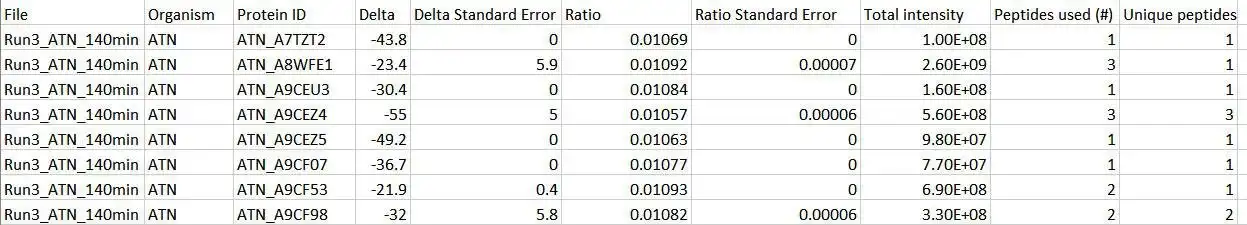
Calis-p (The CALgary approach to ISotopes in proteomics) is a software package to directly extract stable carbon isotope fingerprints (SIFs) of individual species in a microbial community from a metaproteomic dataset. It takes the scored peptide-spectrum match (PSM) tables for samples and the calibration material, and the raw MS data in mzML format as input. In the first step, the software finds the isotopic peaks for each PSM and sums their intensities across a specified retention time window. The identified isotopic patterns (pairs of m/z value + summed intensities) together with the sum formula of the identified peptides are reported and used as the input for the second step of Calis-p to compute delta13C values for all peptides that pass a set of filters, as well as average delta13C values and standard errors for each species. The species delta13C values need to be corrected for instrument isotope fractionation by applying the offset determined using the reference material.Features
- SIF data is directly extracted from a standard metaproteomic dataset
- Efficently and reliably obtain SIF values for a large number of species in a microbial community sample in a high-throughput manner.
This is an application that can also be fetched from https://sourceforge.net/projects/calis-p/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.