This is the Windows app named GenomeRunner to run in Windows online over Linux online whose latest release can be downloaded as GenomeRunner-v4.0-Setup.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named GenomeRunner to run in Windows online over Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
GenomeRunner to run in Windows online over Linux online
DESCRIPTION
Note: This version requires additional SQLite database files. Contact the developers to obtain them.Use http://www.integrativegenomics.org/ for the latest data and analyses.
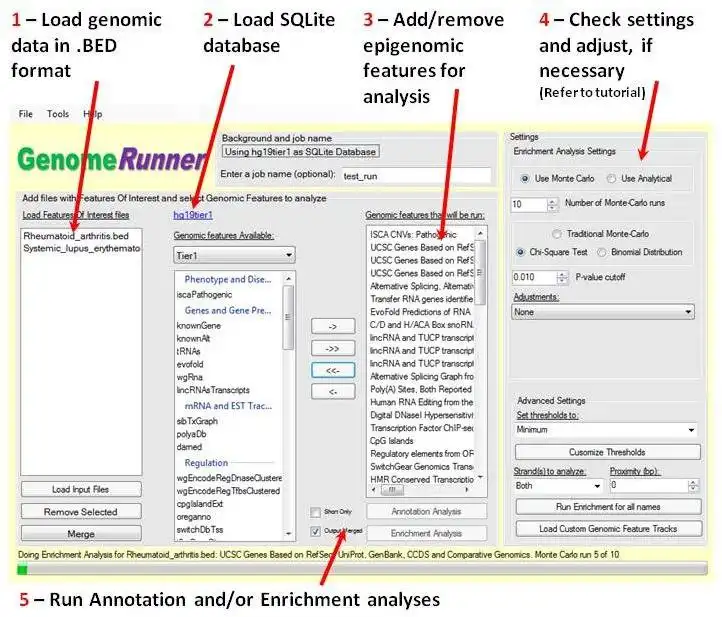
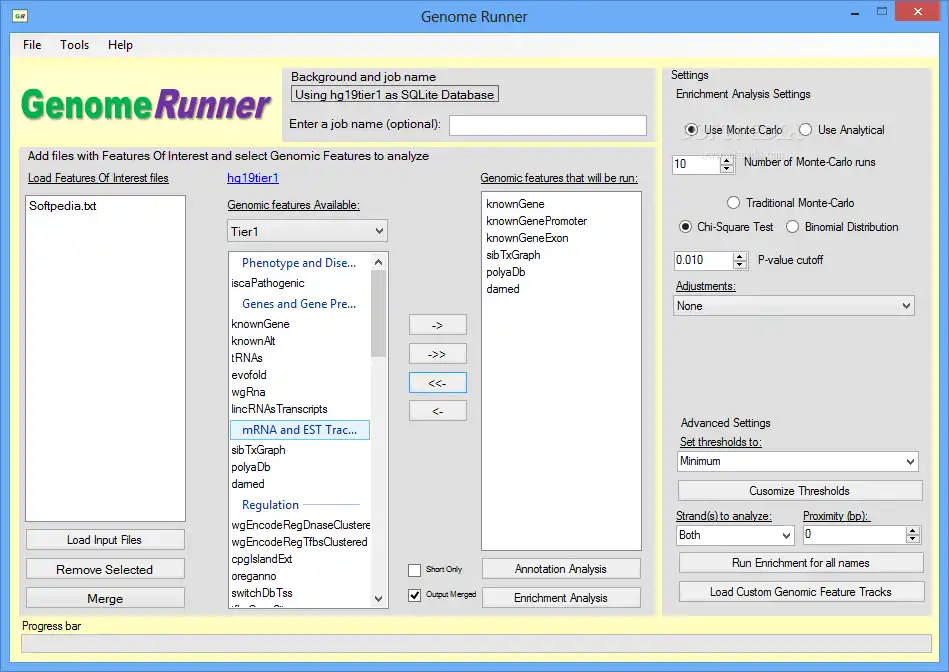
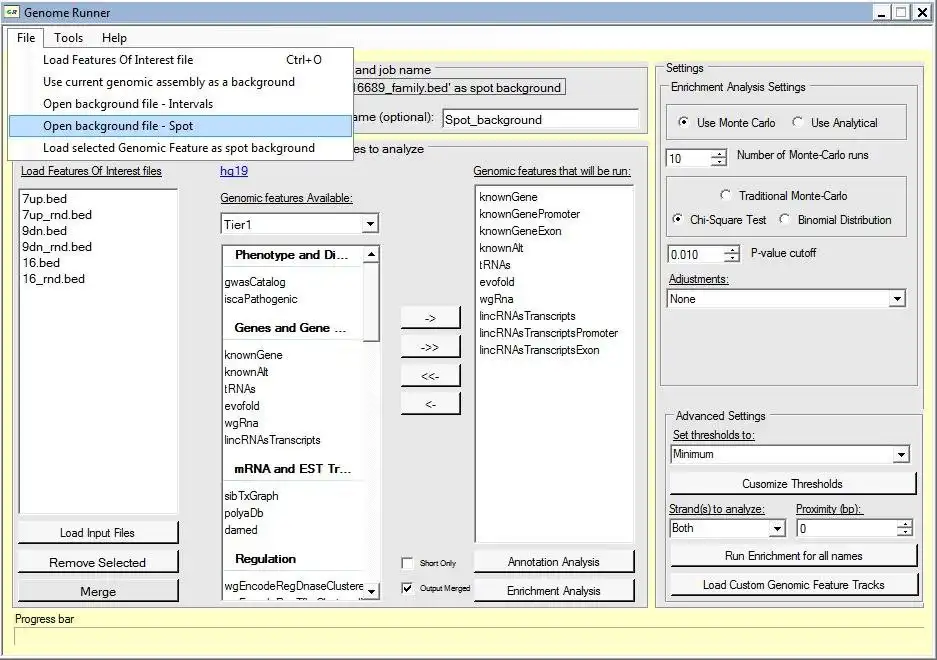
GenomeRunner is a tool for automating genome exploration. It performs annotation and enrichment analyses of user-provided genomic regions (SNPs, ChIP-seq binding sites etc.) against >6,000 (human genome) epigenomic features available from the UCSC genome browser.
Input - any genome-wide data data in .bed format (tab-delimited text file with chrom, chromStart, chromEnd).
Annotation analysis output - detailed annotation of each genomic region in input data. Used to prioritize individual genomic regions by the total number of epigenomic features they co-localize with.
Enrichment analysis output - p-values of statistically significant co-localizations of input genome-wide data with genome annotation features selected for the analysis. Used to prioritize epigenomic features associated with user data.
Features
- Analysis of genomic regions (ChIP-seq, RNA-seq, DNA methylation, SNPs/CNVs etc.)
- ENCODE data
- Annotation and Enrichment analyses
- BED file format (chrom, chromStart, chromEnd)
- Video overview (http://youtu.be/v9p9FClrqXU)
Audience
Science/Research, End Users/Desktop
User interface
Project is a user interface (UI) system
Programming Language
Visual Basic .NET
Database Environment
SQL-based
This is an application that can also be fetched from https://sourceforge.net/projects/genomerunner/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.