This is the Windows app named GMOL whose latest release can be downloaded as gmol_1.1.jar. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named GMOL with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
GMOL
DESCRIPTION
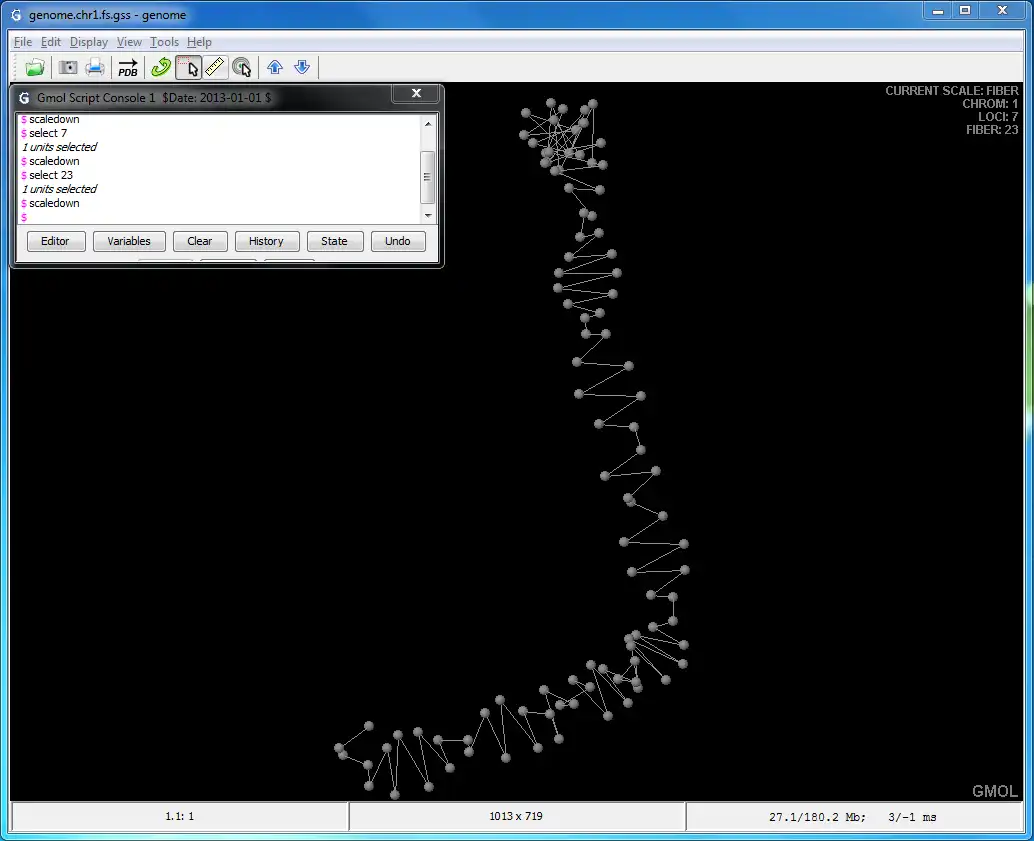
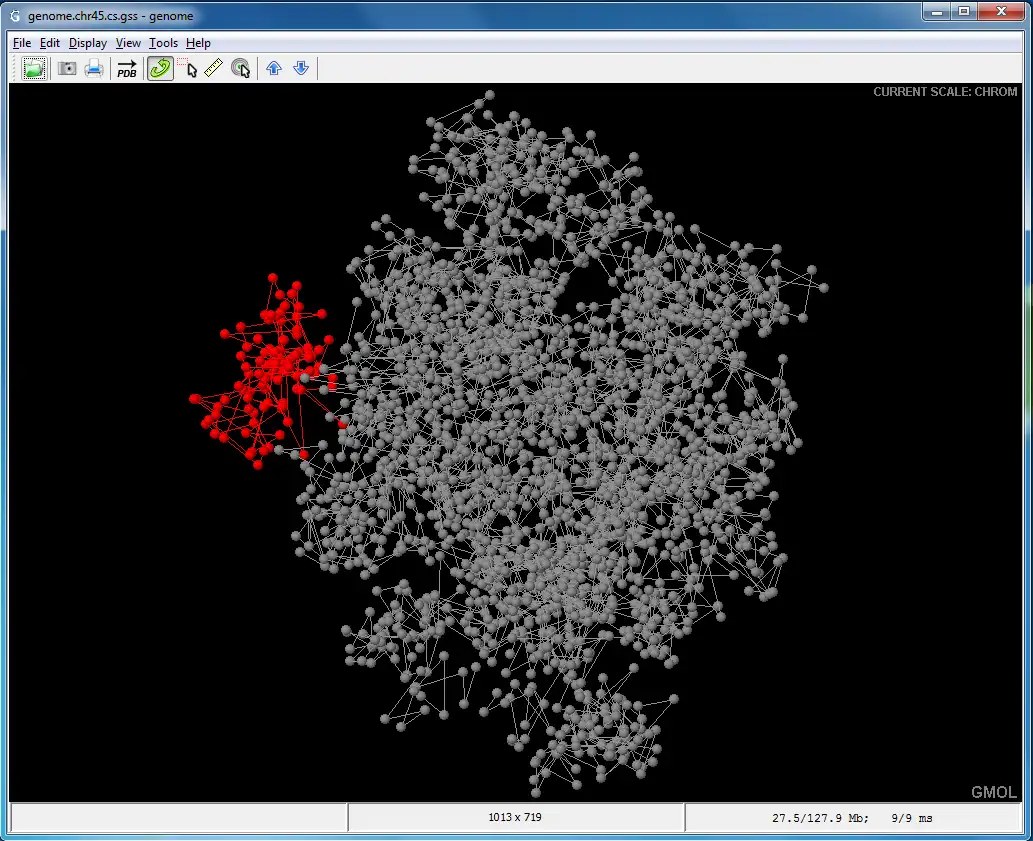
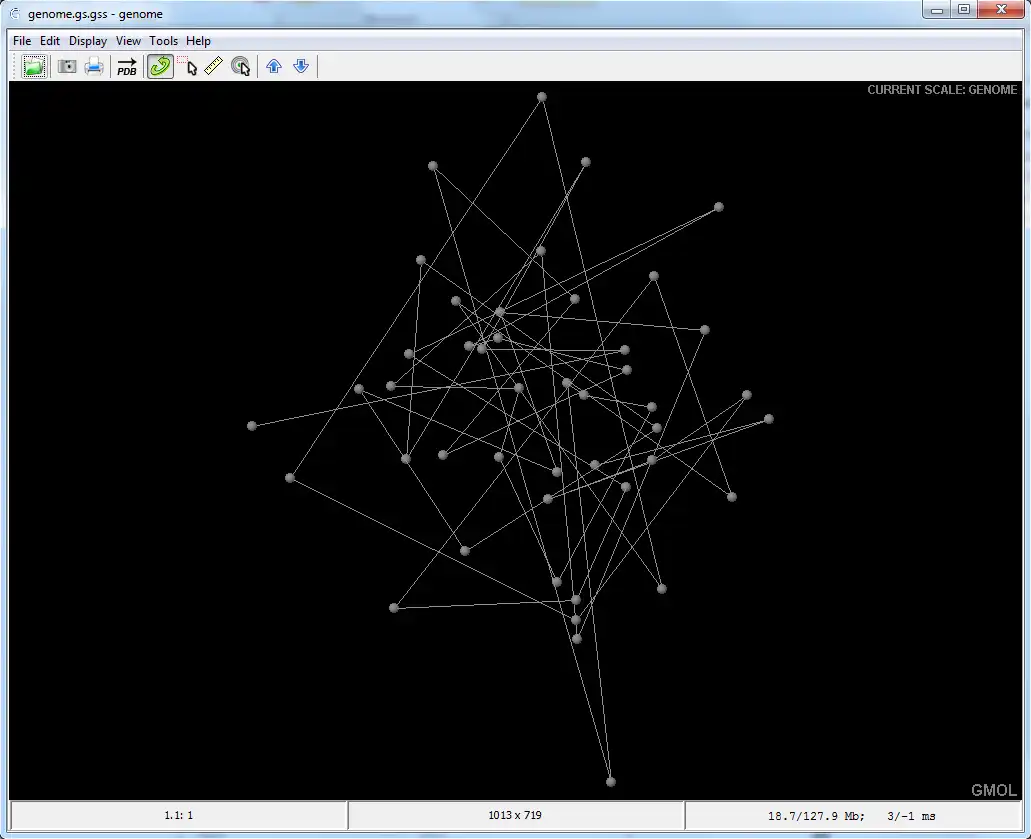
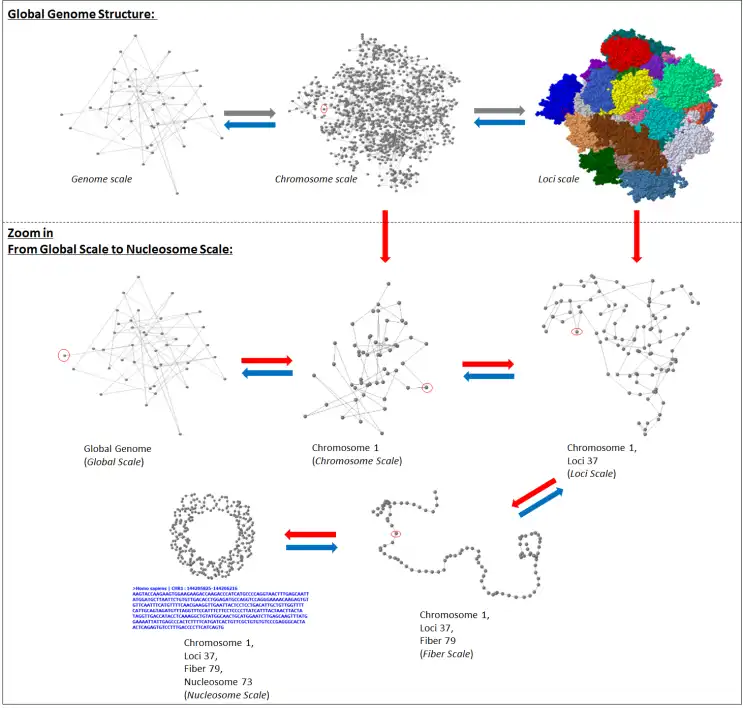
GMOL is an application designed to visualize genome structure in 3D. It allows users to view the genome structure at multiple scales, including: global, chromosome, loci, fiber, nucleosome, and nucleotide. This software was built upon the pre-existing Jmol package by Prof. Cheng's group.The software is developed in Prof. Jianlin Cheng's Bioinformatics, Data Mining and Machine Learning Laboratory in the Computer Science Department at the University of Missouri - Columbia, USA. The project is supported by the National Science Foundation (grant no. DBI1149224).
If you use GMOL in your research, please cite:
Nowotny, Jackson, Avery Wells, Oluwatosin Oluwadare, Lingfei Xu, Renzhi Cao, Tuan Trieu, Chenfeng He, and Jianlin Cheng. "GMOL: an interactive tool for 3D genome structure visualization." Scientific reports 6 (2016): 20802.
Features
- Interactively visualize genome structures in 3D
- Supports multiple scales/resolutions: global, chromosome, loci, fiber, nucleosome, nucleotide
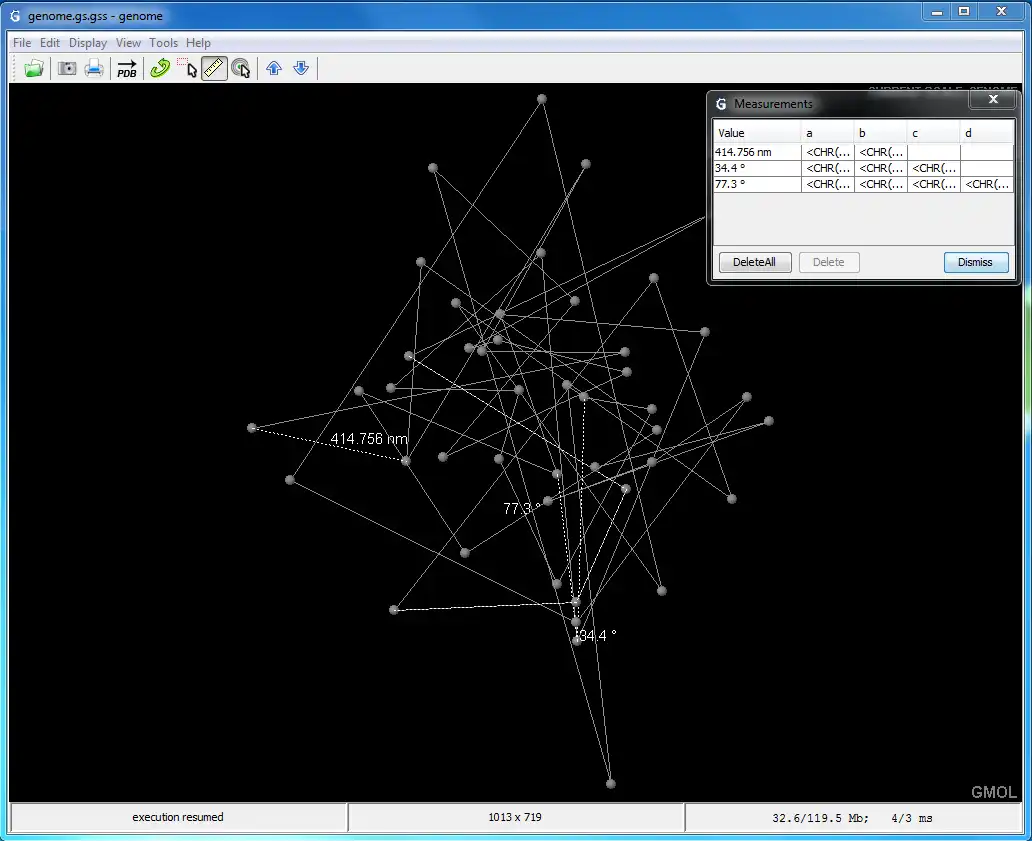
- Measure distances and angles between points in the structure
- Rotate and scale models to analyze them even more
- Select parts of the structure based on index, scale, or sequence information
- Get DNA sequence for selected structures or portions of structures
- Includes existing Jmol functions
Audience
Science/Research, Engineering
Programming Language
Java
This is an application that can also be fetched from https://sourceforge.net/projects/gmol/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.