This is the Windows app named raxmlGUI whose latest release can be downloaded as raxmlGUI1.5b3.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named raxmlGUI with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
raxmlGUI
DESCRIPTION
RELEASE NOTE: Get raxmlGUI 2.0 at the NEW PROJECT LOCATION: https://antonellilab.github.io/raxmlGUI/
raxmlGUI is a graphical user interface to RAxML, one of the most popular and widely used software for phylogenetic inference using maximum likelihood.
A userfriendly graphical front-end for phylogenetic analyses using RAxML (Stamatakis, 2006). Please cite: Silvestro, Michalak (2012) - raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution 12, 335-337. DOI: 10.1007/s13127-011-0056-0
Features
- *NEW VERSION 2.0 NOW AVAILABLE: https://antonellilab.github.io/raxmlGUI/*
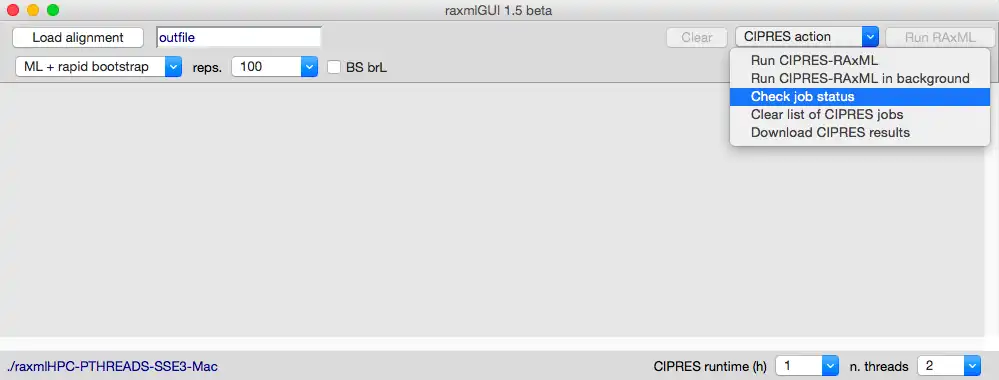
- *version 1.5* adds the possibility to run analyses through CIPRES Science Gateway
- The GUI includes RAxML executables
- Multiplatform compatibility (Mac OS, Windows, Linux)
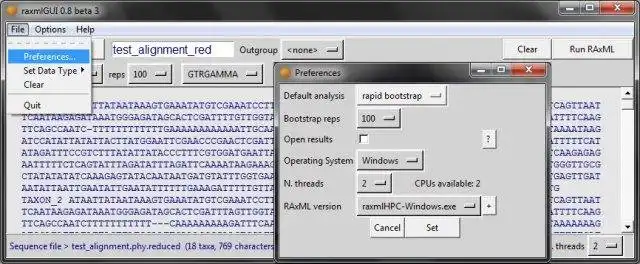
- Requires Python 2.6, 2.7
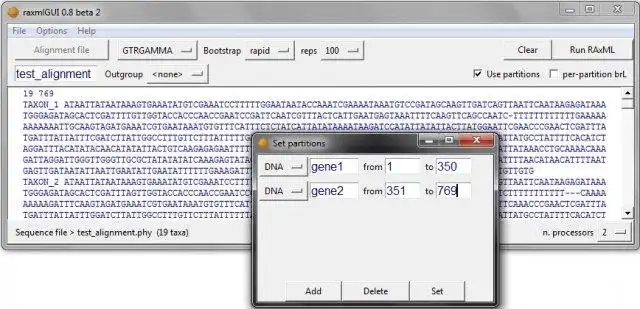
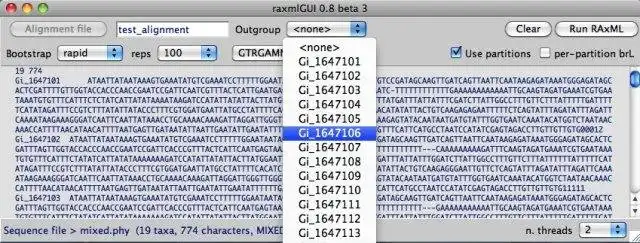
- Intuitive interface for data partitioning, and outgroup selection
- Model selection for DNA, amino acid, binary, and multistate alignments
- Automatic conversion NEXUS to PHYLIP (and vice versa)
- Import FASTA files
- Ancestral state reconstruction
- "Bootstopping" function (Pattengale et al. 2010)
- Interactive exclusion of sites
- Combining multiple alignments with automatic partitioning
- Interactive definition of topological constraints
- Robinson-Foulds pairwise distances
- Per-site likelihoods
Audience
Science/Research
User interface
Tk
Programming Language
Python
Categories
This is an application that can also be fetched from https://sourceforge.net/projects/raxmlgui/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.