This is the Windows app named GenNon-h whose latest release can be downloaded as GenNonH_share.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named GenNon-h with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
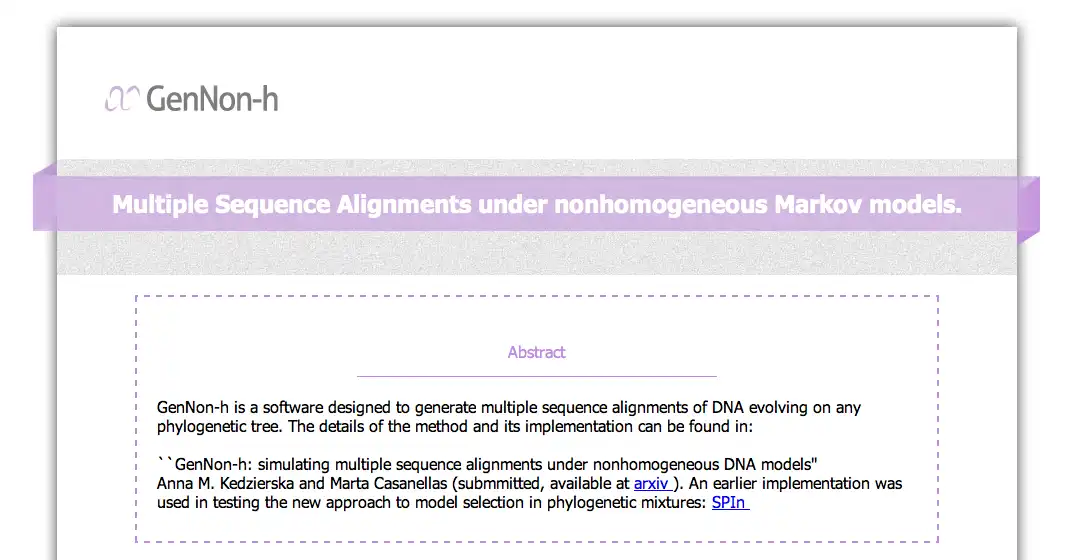
GenNon-h
DESCRIPTION
A number of software packages are available to
generate DNA MSAs evolved under
continuous-time Markov processes on phylogenetic trees. On the other hand,
methods of simulating the DNA MSA directly from the transition matrices
do not exist. Moreover, existing software restricts to
the time-reversible models and it is not optimized to
generate nonhomogeneous data (i.e. placing distinct substitution rates at different lineages).
GenNon-H is the first package designed to generate multiple sequence alignments under
the discrete-time Markov processes on phylogenetic trees, which samples directly from the transition matrices.
Based on the input model and a
phylogenetic tree in the Newick format (with branch lengths measured
as the expected number of substitutions per site), the
algorithm produces DNA alignments of desired length.
GenNon-H is a collaborative project described at http://genome.crg.es/cgi-bin/phylo_mod_sel/AlgGenNonH.pl.
Programming Language
C++
Categories
This is an application that can also be fetched from https://sourceforge.net/projects/gennonh/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.