This is the Windows app named GenOO-HTS whose latest release can be downloaded as genoo-code-2013_04_18.tar.gz. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named GenOO-HTS with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
GenOO-HTS
DESCRIPTION
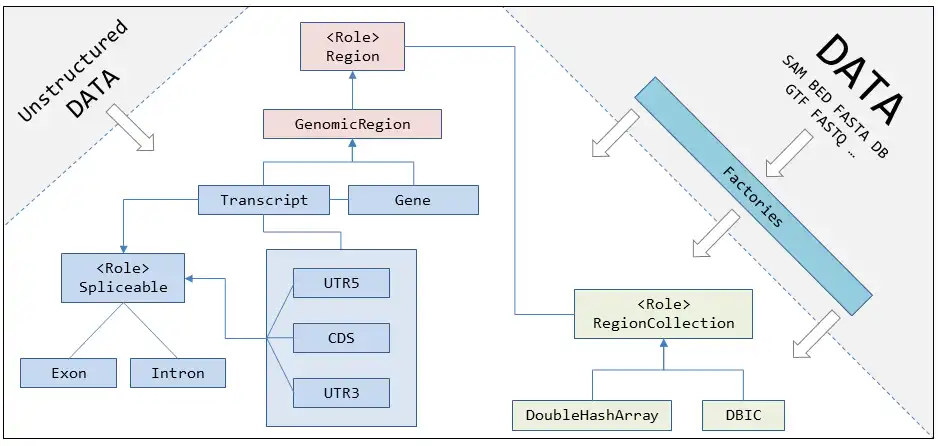
GenOO-HTS [jee-noo] is an open-source; object-oriented Perl framework specifically developed for the design of High Throughput Sequencing (HTS) analysis tools. The primary aim of GenOO-HTS is to make simple HTS analyses easy and complicated analyses possible. GenOO-HTS models biological entities into Perl objects and provides relevant attributes and methods that allow for the manipulation of high throughput sequencing data. Using GenOO-HTS as a core development module reduces the overhead and complexity of managing the data and the biological entities at hand. GenOO-HTS has been designed to be flexible, easily extendable with modular structure and minimal requirements for external tools and libraries.
Features
- Organize biological entities as perl objects (genomic regions, genes, transcripts, introns/exons, etc)
- Organize sequencing entities as perl objects/attributes (sequencing reads, alignments, etc)
- Make I/O from widely used file formats easy (SAM, BED, FASTA, FASTQ)
- Be consistent and easily extendable
Audience
Science/Research, Developers
Programming Language
Perl
Categories
This is an application that can also be fetched from https://sourceforge.net/projects/genoo/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.